-Search query
-Search result
Showing all 33 items for (author: puglisi & ev)

EMDB-17804: 
Bat-Hp-CoV Nsp1 and eIF1 bound to the human 40S small ribosomal subunit
Method: single particle / : Schubert K, Karousis ED, Ban I, Lapointe CP, Leibundgut M, Baeumlin E, Kummerant E, Scaiola A, Schoenhut T, Ziegelmueller J, Puglisi JD, Muehlemann O, Ban N

EMDB-17805: 
MERS-CoV Nsp1 bound to the human 43S pre-initiation complex
Method: single particle / : Schubert K, Karousis ED, Ban I, Lapointe CP, Leibundgut M, Baeumlin E, Kummerant E, Scaiola A, Schoenhut T, Ziegelmueller J, Puglisi JD, Muehlemann O, Ban N

PDB-8ppk: 
Bat-Hp-CoV Nsp1 and eIF1 bound to the human 40S small ribosomal subunit
Method: single particle / : Schubert K, Karousis ED, Ban I, Lapointe CP, Leibundgut M, Baeumlin E, Kummerant E, Scaiola A, Schoenhut T, Ziegelmueller J, Puglisi JD, Muehlemann O, Ban N

PDB-8ppl: 
MERS-CoV Nsp1 bound to the human 43S pre-initiation complex
Method: single particle / : Schubert K, Karousis ED, Ban I, Lapointe CP, Leibundgut M, Baeumlin E, Kummerant E, Scaiola A, Schoenhut T, Ziegelmueller J, Puglisi JD, Muehlemann O, Ban N
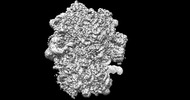
EMDB-26705: 
allo-tRNAUTu1 in the A, P, and E sites of the E. coli ribosome
Method: single particle / : Zhang J, Krahn N, Prabhakar A, Vargas-Rodriguez O, Krupkin M, Fu Z, Acosta-Reyes FJ, Ge X, Choi J, Crnkovic A, Ehrenberg M, Viani Puglisi E, Soll D, Puglisi J
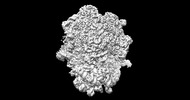
EMDB-26713: 
allo-tRNAUTu1A in the A site of the E. coli ribosome
Method: single particle / : Zhang J, Prabhakar A, Krahn N, Vargas-Rodriguez O, Krupkin M, Fu Z, Acosta-Reyes FJ, Ge X, Choi J, Crnkovic A, Ehrenberg M, Viani Puglisi E, Soll D, Puglisi J

EMDB-26714: 
allo-tRNAUTu1A in the P site of the E. coli ribosome
Method: single particle / : Zhang J, Prabhakar A, Krahn N, Vargas-Rodriguez O, Krupkin M, Fu Z, Acosta-Reyes FJ, Ge X, Choi J, Crnkovic A, Ehrenberg M, Viani Puglisi E, Soll D, Puglisi J

EMDB-26067: 
CryoEM structure of the human 40S small ribosomal subunit in complex with translation initiation factors eIF1A and eIF5B.
Method: single particle / : Lapointe CP, Grosely R, Sokabe M, Alvarado C, Wang J, Montabana E, Villa N, Shin B, Dever T, Fraser C, Fernandez IS, Puglisi JD

PDB-7tql: 
CryoEM structure of the human 40S small ribosomal subunit in complex with translation initiation factors eIF1A and eIF5B.
Method: single particle / : Lapointe CP, Grosely R, Sokabe M, Alvarado C, Wang J, Montabana E, Villa N, Shin B, Dever T, Fraser C, Fernandez IS, Puglisi JD

EMDB-22899: 
Structure of HIV-1 reverse transcriptase initiation complex core
Method: single particle / : Ha B, Larsen KP, Zhang J, Fu Z, Montabana E, Jackson LN, Chen DH, Puglisi EV

EMDB-22900: 
Structure of HIV-1 reverse transcriptase initiation complex core with efavirenz
Method: single particle / : Ha B, Larsen KP, Zhang J, Fu Z, Montabana E, Jackson LN, Chen DH, Puglisi EV

EMDB-22901: 
Structure of HIV-1 reverse transcriptase initiation complex core with nevirapine
Method: single particle / : Ha B, Larsen KP, Zhang J, Fu Z, Montabana E, Jackson LN, Chen DH, Puglisi EV

PDB-7kjv: 
Structure of HIV-1 reverse transcriptase initiation complex core
Method: single particle / : Ha B, Larsen KP, Zhang J, Fu Z, Montabana E, Jackson LN, Chen DH, Puglisi EV

PDB-7kjw: 
Structure of HIV-1 reverse transcriptase initiation complex core with efavirenz
Method: single particle / : Ha B, Larsen KP, Zhang J, Fu Z, Montabana E, Jackson LN, Chen DH, Puglisi EV

PDB-7kjx: 
Structure of HIV-1 reverse transcriptase initiation complex core with nevirapine
Method: single particle / : Ha B, Larsen KP, Zhang J, Fu Z, Montabana E, Jackson LN, Chen DH, Puglisi EV

EMDB-21859: 
CryoEM structure of yeast 80S ribosome with Met-tRNAiMet, eIF5B, and GDP
Method: single particle / : Wang J

PDB-6woo: 
CryoEM structure of yeast 80S ribosome with Met-tRNAiMet, eIF5B, and GDP
Method: single particle / : Wang J, Wang J, Puglisi J, Fernandez IS

EMDB-21582: 
+1 extended HIV-1 reverse transcriptase initiation complex core (pre-translocation state)
Method: single particle / : Larsen KP, Jackson LN

EMDB-21583: 
+3 extended HIV-1 reverse transcriptase initiation complex core (pre-translocation state)
Method: single particle / : Larsen KP, Jackson LN

EMDB-21584: 
+3 extended HIV-1 reverse transcriptase initiation complex core (intermediate state)
Method: single particle / : Larsen KP, Jackson LN

EMDB-21585: 
+3 extended HIV-1 reverse transcriptase initiation complex core (displaced state)
Method: single particle / : Larsen KP, Jackson LN

EMDB-22002: 
+1 extended HIV-1 reverse transcriptase initiation complex (pre-translocation state, global map)
Method: single particle / : Larsen KP, Jackson LN, Zhang J, Puglisi EV

EMDB-22003: 
+3 extended HIV-1 reverse transcriptase initiation complex (pre-translocation state, unmasked map)
Method: single particle / : Larsen KP, Jackson LN, Zhang J, Puglisi EV

EMDB-22004: 
+3 extended HIV-1 reverse transcriptase initiation complex (intermediate state, unmasked map)
Method: single particle / : Larsen KP, Jackson LN, Zhang J, Puglisi EV

EMDB-22005: 
+3 extended HIV-1 reverse transcriptase initiation complex (displaced state, unmasked map)
Method: single particle / : Larsen KP, Jackson LN, Zhang J, Puglisi EV

PDB-6waz: 
+1 extended HIV-1 reverse transcriptase initiation complex core (pre-translocation state)
Method: single particle / : Larsen KP, Jackson LN, Kappel K, Zhang J, Puglisi EV

PDB-6wb0: 
+3 extended HIV-1 reverse transcriptase initiation complex core (pre-translocation state)
Method: single particle / : Larsen KP, Jackson LN, Kappel K, Zhang J, Puglisi EV

PDB-6wb1: 
+3 extended HIV-1 reverse transcriptase initiation complex core (intermediate state)
Method: single particle / : Larsen KP, Jackson LN, Kappel K, Zhang J, Chen DH, Puglisi EV

PDB-6wb2: 
+3 extended HIV-1 reverse transcriptase initiation complex core (displaced state)
Method: single particle / : Larsen KP, Jackson LN, Kappel K, Zhang J, Chen DH, Puglisi EV

EMDB-7032: 
Global architecture of the HIV-1 reverse transcriptase initiation complex
Method: single particle / : Larsen KP, Mathiharan YK, Chen DH, Puglisi JD, Skiniotis G, Puglisi EV

EMDB-7540: 
Cryo-EM map of an HIV-1 reverse transcriptase initiation complex in magnesium chloride imaging buffer
Method: single particle / : Larsen KP, Chen DH, Puglisi JD, Puglisi EV

EMDB-7031: 
Architecture of HIV-1 reverse transcriptase initiation complex core
Method: single particle / : Larsen KP, Mathiharan YK, Chen DH, Puglisi JD, Skiniotis G, Puglisi EV

PDB-6b19: 
Architecture of HIV-1 reverse transcriptase initiation complex core
Method: single particle / : Larsen KP, Mathiharan YK, Chen DH, Puglisi JD, Skiniotis G, Puglisi EV
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
